
Welcome to website for the Genomics Aotearoa ONT MinION workshop, which ran at the University of Otago, Dunedin, from 17th–19th April, 2018.
Schedule
Presentations
- Simone Cree:
- Chris Winefield:
- Benjamin Schwessinger:
- Louise Judd:
- Kirston Barton:
- HMW DNA on Nanopore
- Lab session: QCing your DNA before a library prep
- Alexis Lucattini:
Tutorials
- Handling MinION data from your laptop
- Setting up a virtual environment
- Handling fast5 data on a server
Information for Participants
Sequencing results
- Here are some stats for each run, produced by Rob Lanfear’s MinIONQC.R script.
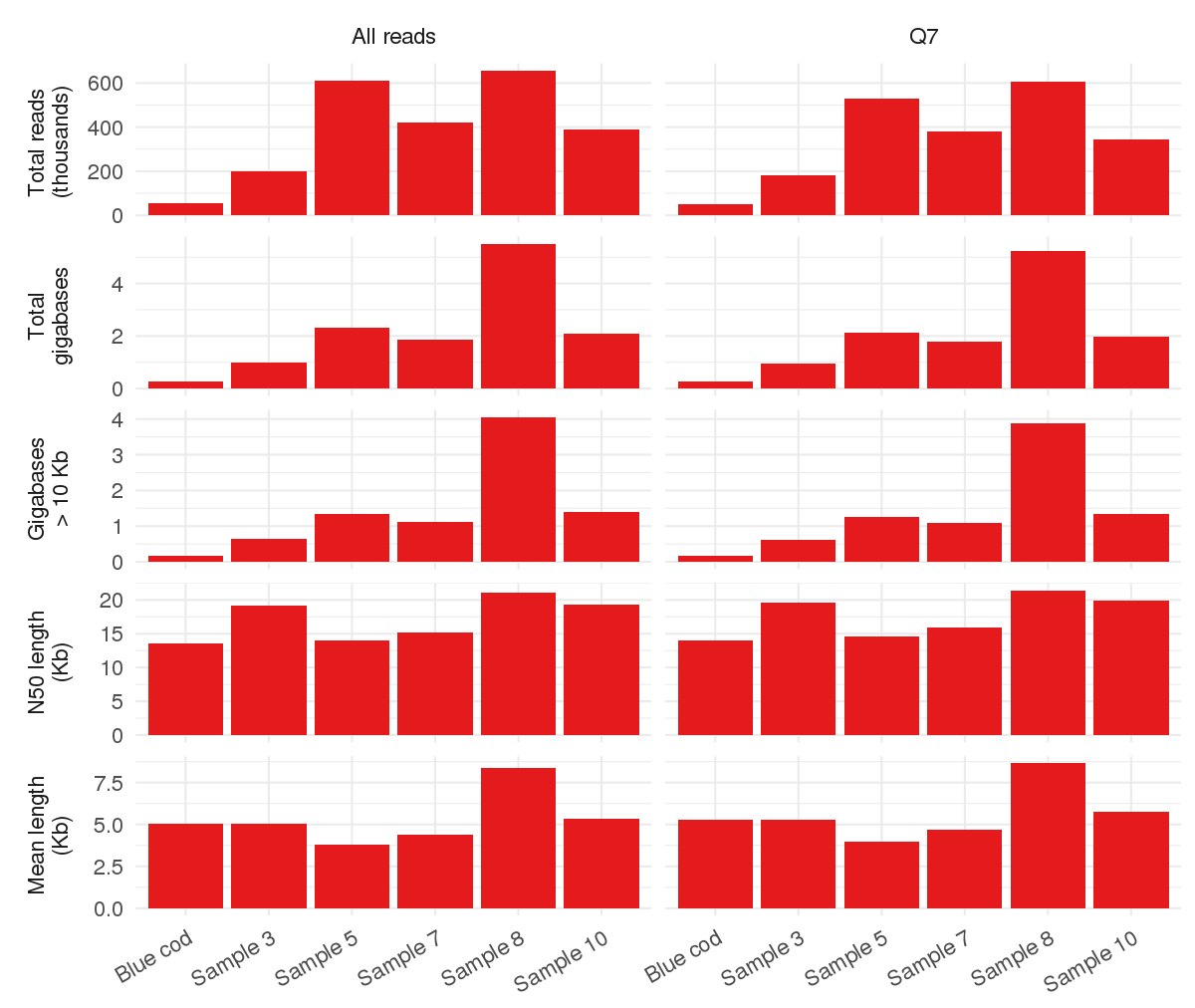
-
There’s a
.tar.gzfile here with all the output from the script. The meaning of each plot is explained on the github repo. -
Here’s the weighted read length histogram for each sample.
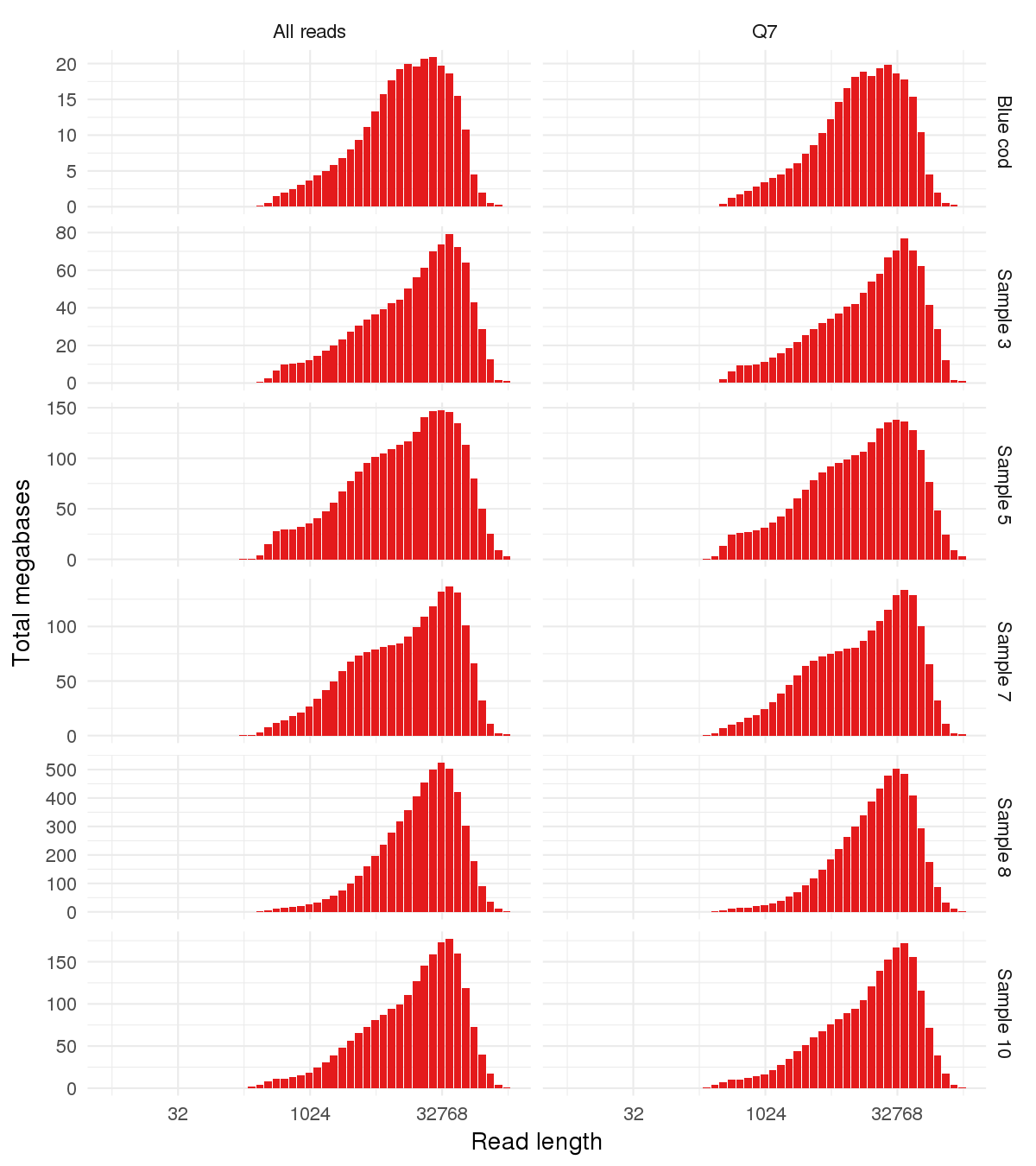
- You can also find the sample QC and library yield results at the following links, to compare to the sequencing results:
Links
- Nanopore group on protocols.io
- Insect HMW DNA protocol used to extract samples 1–10
- Olin’s
ultra cheapreasonably priced magnetic rack